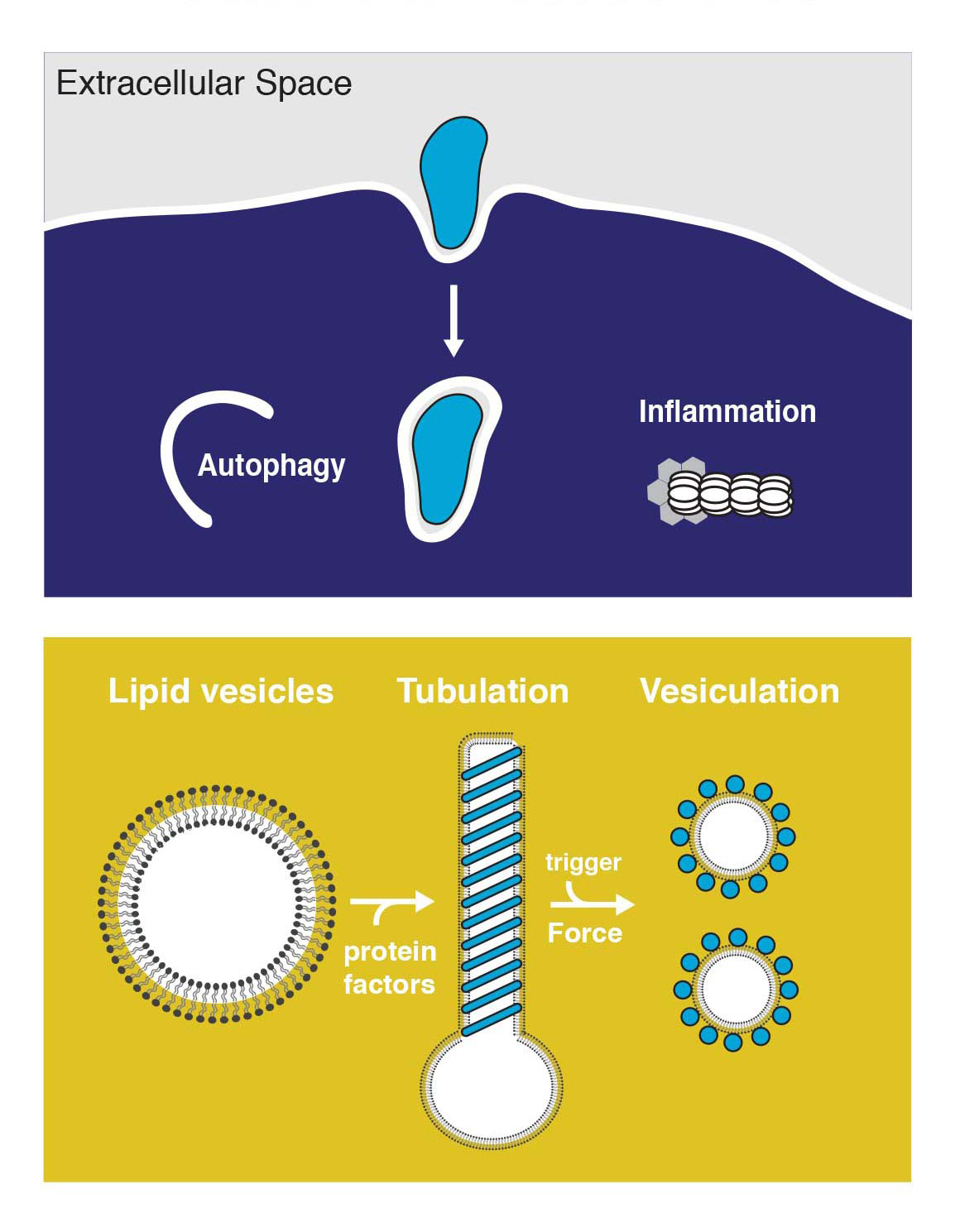
The ability of most cell lineages to defend themselves individually against infection can be considered as the most ancestral form of defense against pathogen pressure. We are interested in the intriguing interplay of autophagy and intracellular bacteria, where the host employs autophagy for bacterial clearance while some pathogens re-direct autophagy as a pro-survival strategy. We work towards understanding the mechanisms of restricting intracellular pathogens by studying three-dimensional structures of the protein machinery involved in this form of cytosolic host defense.
Intracellular trafficking is characterized by dynamic membrane remodeling events. Pathogens hijack autophagy and other intracellular membranes to subvert immune recogonition, replicate and to escape from host cells. We currently lack a detailed view of how the pathogen protein machinery assembles on membranes to remodel its replicative compartments. The overarching goal of this research program is to understand the structures and mechanism of these protein machineries, and those of the host involved in restricting intracellular pathogens. We use nanotechnology alongside biochemistry and cell biology to build reconstituted systems that we can manipulate precisely, and we use electron imaging methods to visualize them in molecular detail.
Immunity-related large GTPases
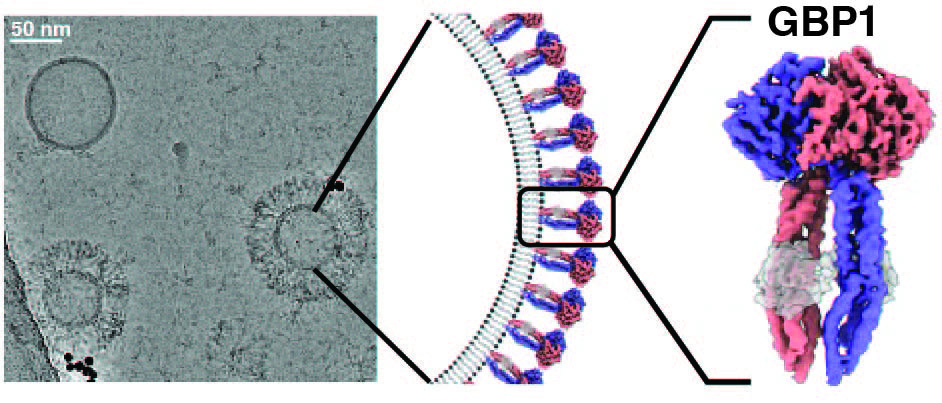 GBPs are interferon-inducible guanosine triphosphate hydrolases (GTPases) that promote elimination of intracellular pathogens. Their antimicrobial activity depends on the formation of homo- and heterooligomeric complexes on the membrane of pathogen-associated compartments or cytosol invasive bacteria, and requires GTPase activity. How GTP hydrolysis primes GBPs for coatomer formation, and how this mobilise bacterial membrane components for effector activation remains unclear. We use a combination of in vitro and in situ electron cryo-tomography (cryo-ET), high-resolution fluorescence microscopy and single-molecule biophysics to address these questions.
GBPs are interferon-inducible guanosine triphosphate hydrolases (GTPases) that promote elimination of intracellular pathogens. Their antimicrobial activity depends on the formation of homo- and heterooligomeric complexes on the membrane of pathogen-associated compartments or cytosol invasive bacteria, and requires GTPase activity. How GTP hydrolysis primes GBPs for coatomer formation, and how this mobilise bacterial membrane components for effector activation remains unclear. We use a combination of in vitro and in situ electron cryo-tomography (cryo-ET), high-resolution fluorescence microscopy and single-molecule biophysics to address these questions.
We could recently show how nucleotide binding promotes large-scale conformational changes in GBP1 that expose the isoprenylated carboxyl-terminus for association with membranes. This conformation is critical for coatomer formation and permits intercalation into the lipopolysaccharide layer on the outer membrane of gram-negative bacterial pathogens. These data form only the beginning of our mechanistic understanding of how GBPs bring about their immune effector function. While focussing on answering these biological questions at the molecular level, we are investing into technological developments for correlative molecular imaging that are required to bring us closer to this goal.
Membrane remodelling by Chlamydia trachomatis
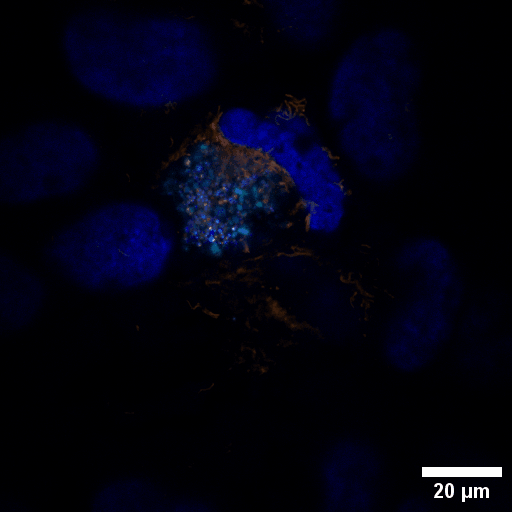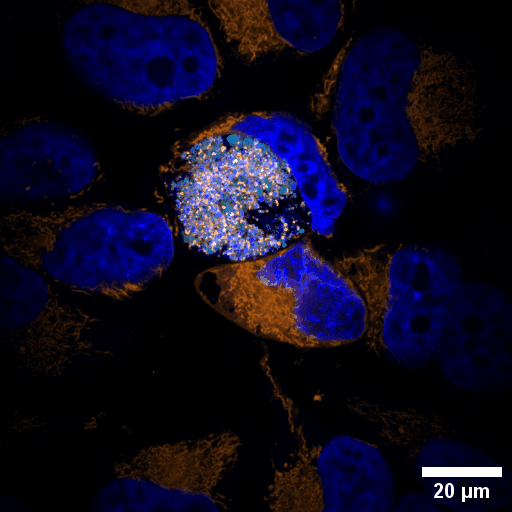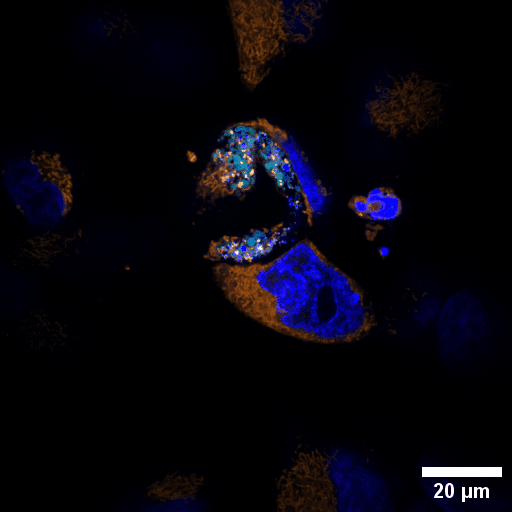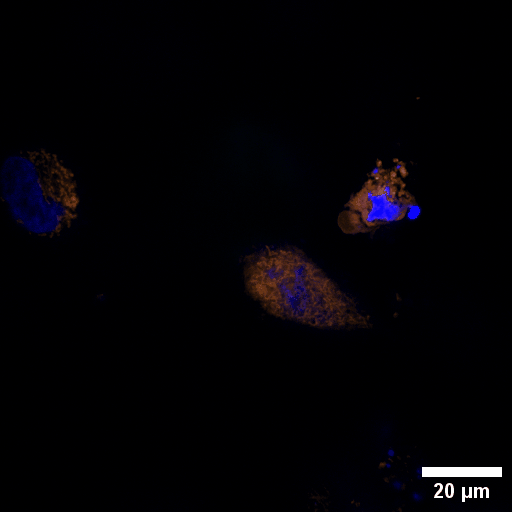 Chlamydia trachomatis (Ctr) is an obligate intracellular human pathogen and the leading infectious cause of blindness. It also is the most frequent sexually transmitted bacterial infection linked to adverse pregnancy outcomes and infertility. Once it enters the cell, the pathogen drives the conversion of the endocytic compartment into a replicative niche termed inclusion. Chlamydia is exceptionally dependent on its host cell because of the particularities of their developmental biology. Aside from rewiring cellular metabolic pathways Ctr infection also triggers large-scale remodelling of intracellular membranes along the progression through its infectious cycle. We are using correlative light and electron microscopy imaging to characterise these processes at the ultrastructural and molecular level.
Chlamydia trachomatis (Ctr) is an obligate intracellular human pathogen and the leading infectious cause of blindness. It also is the most frequent sexually transmitted bacterial infection linked to adverse pregnancy outcomes and infertility. Once it enters the cell, the pathogen drives the conversion of the endocytic compartment into a replicative niche termed inclusion. Chlamydia is exceptionally dependent on its host cell because of the particularities of their developmental biology. Aside from rewiring cellular metabolic pathways Ctr infection also triggers large-scale remodelling of intracellular membranes along the progression through its infectious cycle. We are using correlative light and electron microscopy imaging to characterise these processes at the ultrastructural and molecular level.
Structural basis for membrane targeting and coatomer formation by human GBP1
Kuhm T, Taisne CM, de Agrela Pinto C, Gross L, Giannopolou EA, Huber ST, Pardon E, Steyaert J, Tans S, Jakobi AJ
bioRxiv 2023.03.28.534355
Structural basis of p62/SQSTM1 helical filaments and their role in cellular cargo uptake
Jakobi AJ, Huber ST, Mortensen SA, Schultz SW, Palara A, Kuhm T, Lamark T, Johansen T, Brech A, Sachse C
Nature Communications 11:440 (2020)
We work on a number of large nanomachines involved in bacterial and mammalian cell biology. A major focus of current efforts are gas vesicles, a fascinating microbial motility system imparting buyoancy on aquatic bacteria and archea. We have been able to determine the detailed atomic structure of the gas vesicle wall, formed by a 7 kDa protein GvpA that assembles into a selectively permeable proteinaceous shell that can grow hundreds of nanometers in length and encloses a gas-filled volume. We have also grown accustomed to the fact that many of the systems we choose to work on miraculously appear to form helical assemblies. We therefore have broad experience and a keen interest in cryo-EM structure determination of helical assemblies.
Microbial gas vesicles
Gas vesicles are intracellular, gas-filled nanostructures that allow prokaryotes to float vertically to the surface of their aqueous habitat. Gas vesicles are unique among prokaryotic motility systems in that their structure alone imparts the function of motility. Aquatic cyanobacteria, for example, utilise this form of motility to optimise light harvesting conditions for photosynthesis by rising to the surface. Massive accumulation of such cyanobacterial aggregates can be seen in freshwater lakes, colloquially called algal blooms. We are deeply intrigued by the molecular architecture of this prokaryotic motility machinery and are using a combination of structural and biophysical approaches to understand their unique physical properties and the mechanism of gas vesicle assembly and morphogenesis.
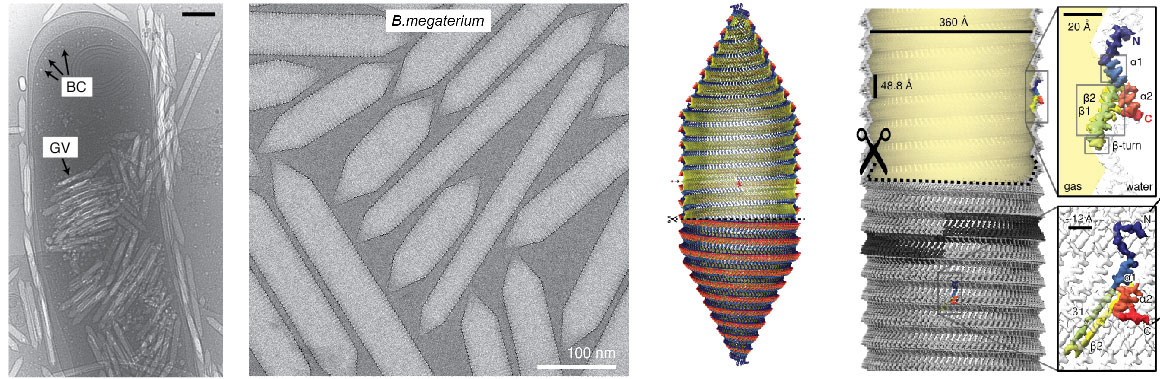
Cryo-EM structure of gas vesicles for buoyancy-controlled motility
Huber ST, Terwiel D, Evers W, Maresca D, Jakobi AJ
Cell 186:975-98 (2023)
Structural biology of gas vesicles: Historical milestones and current knowledge
Huber ST*, Jakobi AJ*
Biochem Soc Trans 186:975-98 (2024)
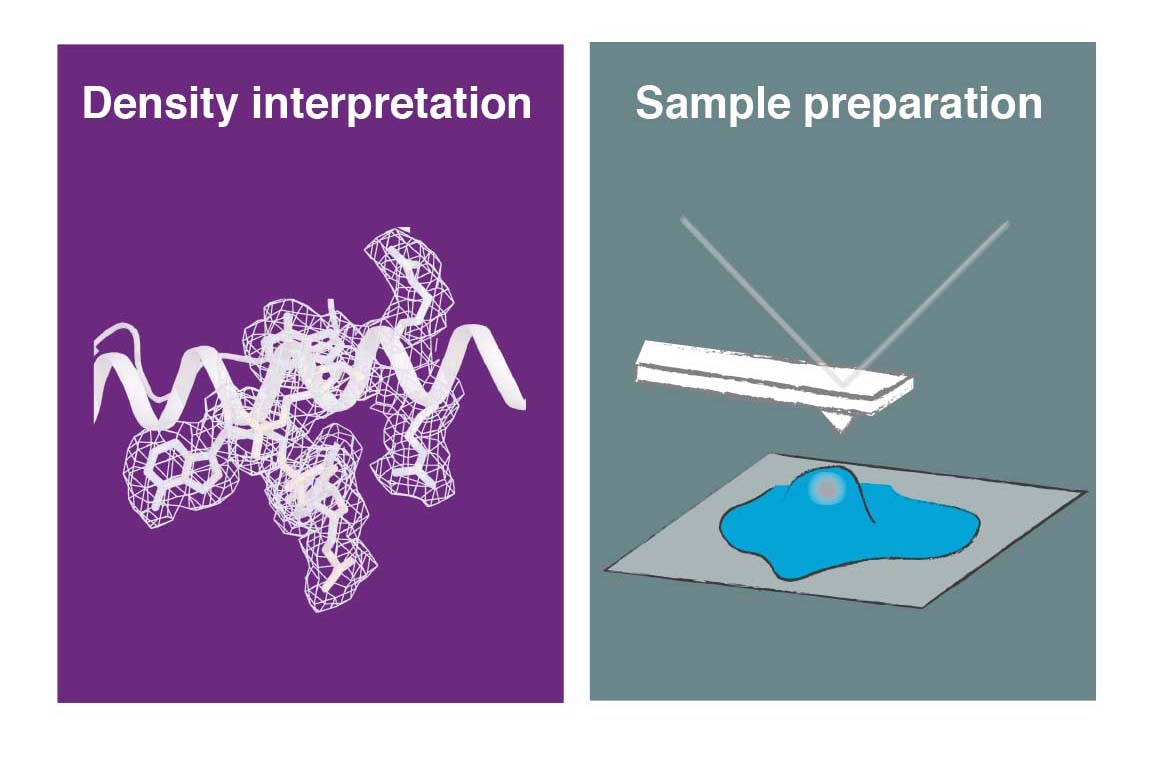
Another major area of our research is the development and application of new methods for cryo-EM. We aim to facilitate some of the current experimental challenges, such as sample preparation for single-particle analysis and in situ cryo-ET to improve the overall performance and efficiency of the technique and to broaden it applicability to a wider base of non-expert users. This interest also extends to computational method development, in particular to efficient methods to simulate and evaluate structural heterogeneity in cryo-EM/ET data, to aid the estimation of helical symmetry paramters and to facilitating the last step of cryo-EM structure determination, which is the interpretation of the derived 3D maps in terms of an atomic model.
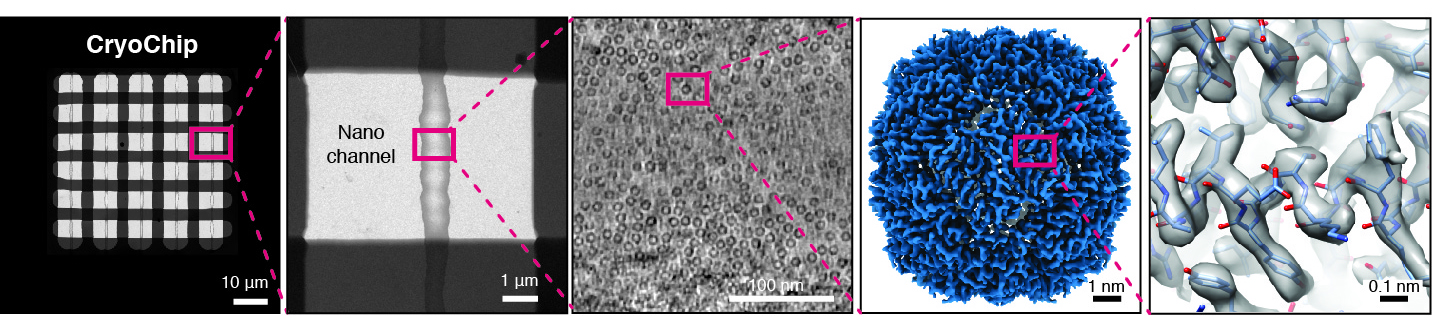
Nanofabricated MEMS supports for cryo-EM sample preparation
We are developing a novel generation of cryo-EM imaging support based on Micro-/nanoelectromechanical Systems (M/NEMS) technology. M/NEMS nanofabrication techniques employed provide opportunities to miniaturise and automate cryo-EM sample preparation. Together with our partner SmartTip we have demonstrated that nanofluidic cavities with well-defined geometry can be used to prepare cryo-EM specimens with uniform ice thickness from picoliter sample volumes. Several key challenges remain to be addressed in order to transform this approach into a viable alternative to widely used holey support films. We are working towards solutions to these challenges and also exploring new application areas of MEMS-based cryo-EM sample preparation such as time-resolved experiments and correlative imaging.
Facilitating sample preparation and correlative imaging workflows for in situ cryo-ET/CLEM
Cryogenic focused ion beam (cryo-FIB) micromachining is used to prepare a thin lamella-shaped sample out of a frozen-hydrated cell for cryo-ET imaging. Current workflows for targeted cryo-FIB milling require multiple sample transfers prone to contamination and relocation/registration errors. In close collaboration with Jacob Hoogenboom, we are working toward improved workflows for in situ fluorescence microscopy-guided FIB fabrication of a frozen-hydrated lamella. This includes development of a coincident three-beam cryogenic correlative microscope and 4D STEM approaches to allow monitoring both lamella thickness and amorphous state during lamella production. The ability to check the lamella during and after the milling process results in a higher success rate in the fabrication process and will increase the throughput of fabrication for lamellae suitable for high-resolution imaging.
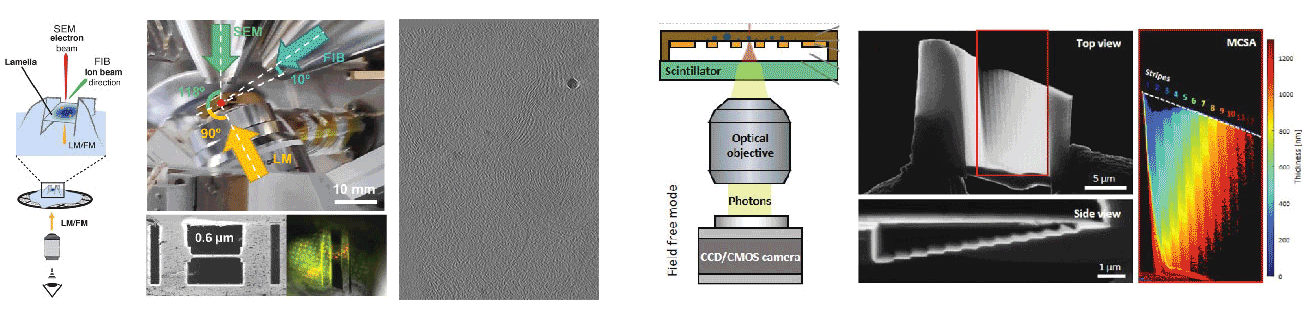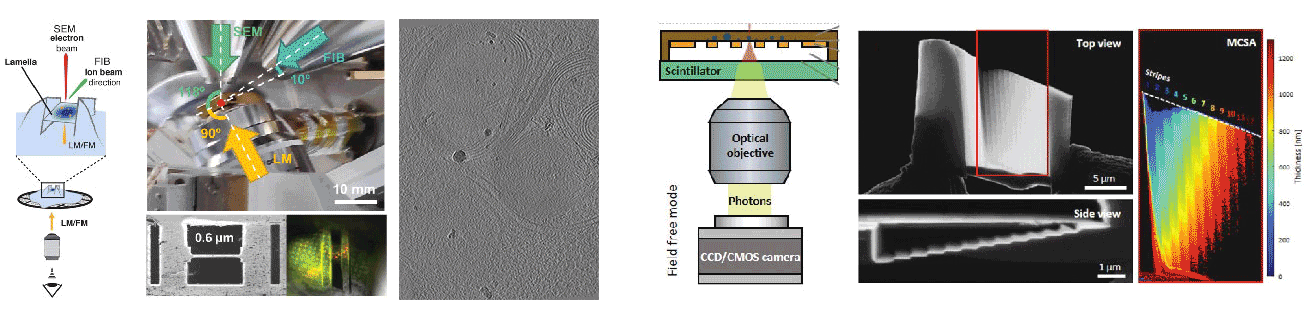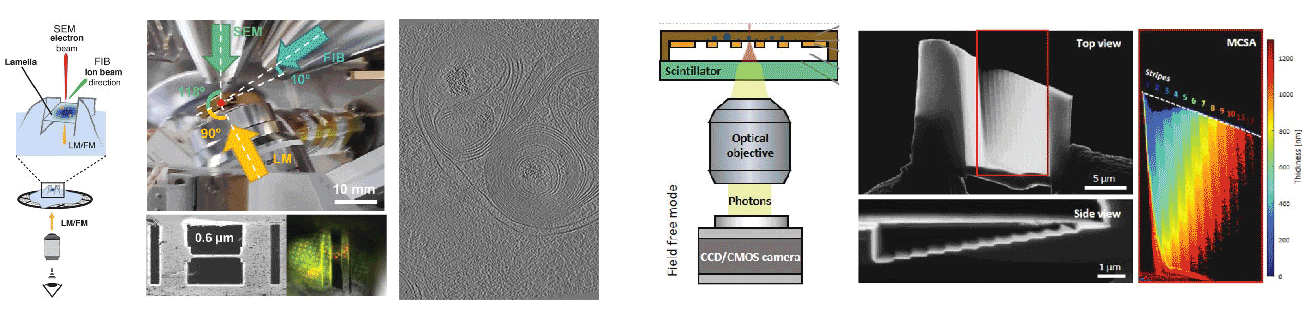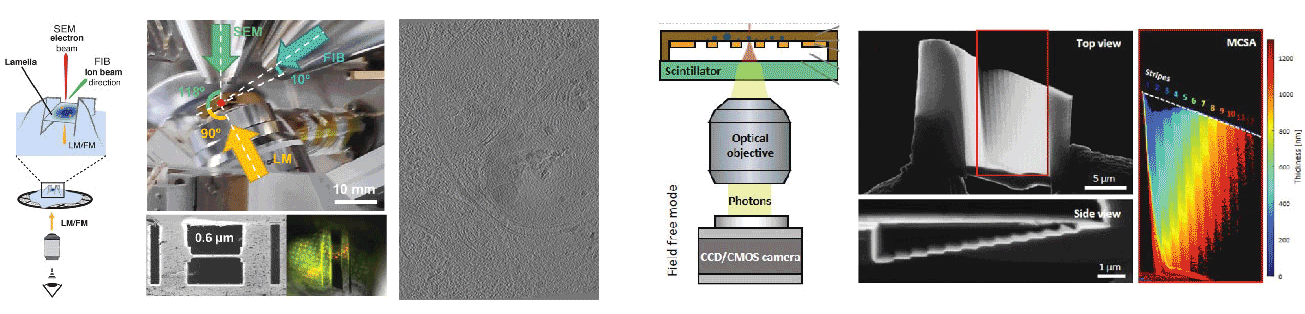
A cryogenic, coincident fluorescence, electron, and ion beam microscope
Boltje DB, Hoogenboom JP, Jakobi AJ, Jensen GJ, Jonker CTH, Kaag MJ, Koster AJ, Last MGF, de Agrela Pinto C, Plitzko JM, Raunser S, Tacke S, Wang Z, van der Wee EB, Wepf R, ten Hoedt S
Elife 11:e82891 (2022)
Robust Local Thickness Estimation of Sub-Micrometer Specimen by 4D-STEM
Skoupý R, Boltje DB, Slouf M, Mrázová K, Láznička T, Taisne CM, Krzyzanek*, Hoogenboom JP*, Jakobi AJ*
Small Methods 7:2300258 (2023)
Density interpretation and model building
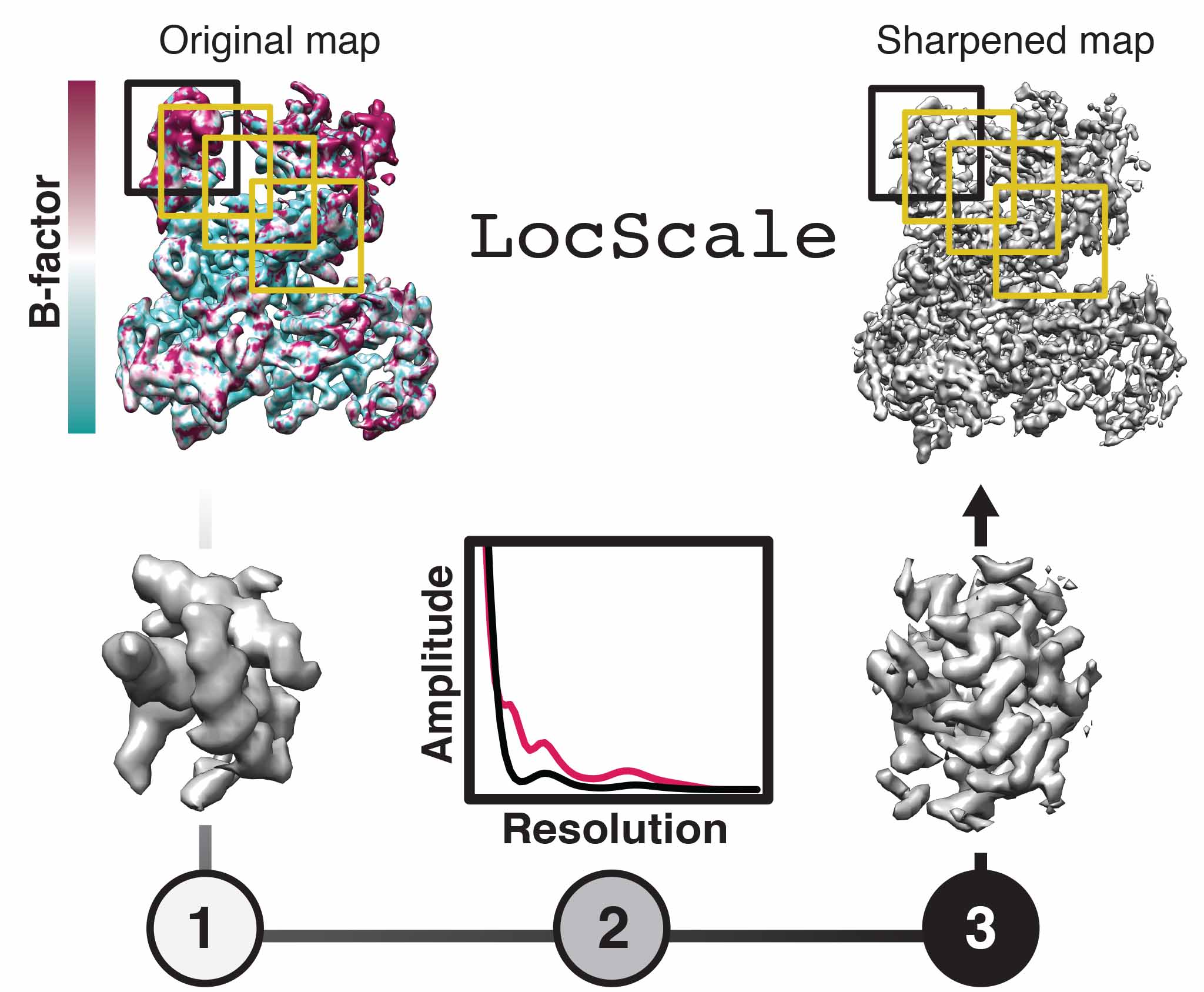 We develop methods to facilitate interpretation of cryo-EM density maps and improve the accuracy of models derived from them. We have introduced the concept of local sharpening to optimize contrast in maps with local resolution variation and adaptive restraint weigthing for atomic model refinement against a single map target. Our efforts in improving density interpretation methods are continued as part of the CCP-EM project.
We develop methods to facilitate interpretation of cryo-EM density maps and improve the accuracy of models derived from them. We have introduced the concept of local sharpening to optimize contrast in maps with local resolution variation and adaptive restraint weigthing for atomic model refinement against a single map target. Our efforts in improving density interpretation methods are continued as part of the CCP-EM project.
Model-based local density sharpening of cryo-EM maps
Jakobi AJ, Wilmanns M, Sachse C
Elife 6:e27131 (2017)
Electron scattering properties of biological macromolecules and their use for cryo-EM map sharpening
Bharadwaj A, Jakobi AJ
Faraday Discuss. 240:168-183 (2022)




